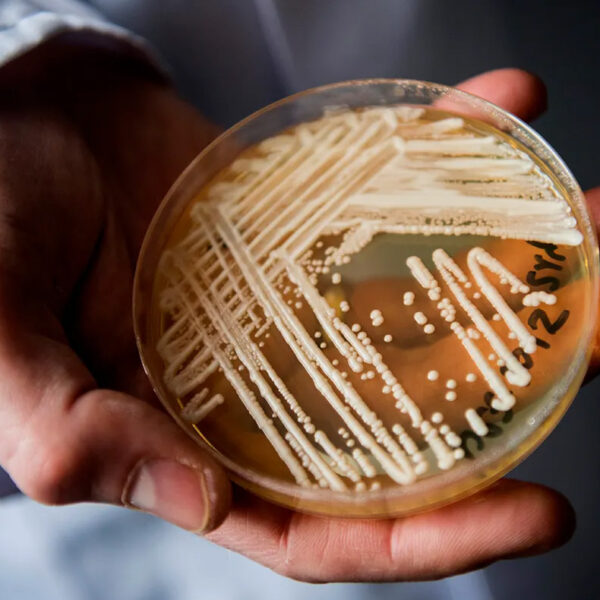
Bioinformatics
The Hybrid Science
The potential of science in a data-driven world just keeps growing, especially in the bioinformatics field.
Data and analytics applied to science give rise to more rapid discoveries, swifter technological advances, and most importantly, more immediate impacts on the human condition.
Integrated knowledge from biological, computational and mathematical disciplines creates a unique professional perspective preparing graduates to play pivotal roles in today’s cutting-edge life sciences, biotechnology and pharmaceutical industries.
Students take classes in a range of disciplines including molecular biology, biochemistry, statistics, ethics, data mining and machine learning, giving them a competitive edge in the ever-expanding life sciences sector.
This is so much more than crunching numbers. It’s changing lives.
- Bioinformatics Enterprise
- Biotechnology
- Data Analytics
- Health Informatics
- Medical Health Informatics
- Omics
5-9% expected national job growth for bioinformatic scientists by 2026 (US Department of Labor)
AVERAGE ANNUAL SALARY FOR BIOINFORMATICIANS IN 2018 (GLASSDOOR)
Faculty and Support Staff
Jared Auclair
Jennifer Darvesh
Diversity & Inclusion
The College of Science supports a culture where each person feels they belong, regardless of race, color, religion, religious creed, genetic information, sex, gender, gender identity, sexual orientation, age, national origin, ancestry, veteran or disability status. We celebrate the diversity of our community, and we seek to expand representation to further excellence. We commit to be a College where members act with respect, trust, collaboration, and communication, and where inappropriate behavior is reported and acted on without fear of retaliation.

Introduces the concepts of probability and statistics used in bioinformatics applications, particularly the analysis of microarray data. Uses statistical computation using the open-source R program. Topics include maximum likelihood; Monte Carlo simulations; false discovery rate adjustment; nonparametric methods, including bootstrap and permutation tests; correlation, regression, ANOVA, and generalized linear models; preprocessing of microarray data and gene filtering; visualization of multivariate data; and machine-learning techniques, such as clustering, principal components analysis, support vector machine, neural networks, and regression tree.
Covers various aspects of data mining, including classification, prediction, ensemble methods, association rules, sequence mining, and cluster analysis. The class project involves hands-on practice of mining useful knowledge from a large data set.
Intended for those familiar with the basics of genetics, molecular and cellular biology, and biochemistry, all of which are required to appreciate the beauty, power, and importance of modern genomic approaches. Introduces the latest sequencing methods, array technology, genomic databases, whole genome analysis, functional genomics, and more.
News