Proteins are one of the most essential parts of any living organism. They provide cellular structure, carry out a variety of functions, and work to regulate tissues and organs. In order to have proteins, however, systems rely on the ribosome – a biological machine within the cell that makes protein.

Paul Whitford, Assistant Professor of Theoretical Condensed Matter and Biological Physics
Northeastern Assistant Professor of Physics Paul Whitford has been focusing his research on the essential and complex functional movements of the ribosome for the past several years. Since receiving his PhD, he has been looking at protein folding and conformational changes, as well as developing a class of models that can be applied to a broad range of biomolecules, which he continues to use today. His recent studies have followed two parallel tracks that are just the beginning of many exciting discoveries to come.
The ribosome works to produce proteins in the cell. In order to do this, it undergoes a sequence of conformational rearrangements, which it must go through quickly and repeatedly. To produce proteins, the ribosome must decode the cell’s genetic message with the help of tRNA molecules (transfer RNAs, which decode messenger RNA sequences into proteins at specific sites of the ribosome). If the ribosome does not recruit the correct tRNA molecule, the produced protein will have errors, which can have disastrous effects on the cell. This means that the ribosome must work accurately, as well as quickly and efficiently, in order to ensure that the cell will continue to grow without wasting precious energy.
Whitford’s first project studied how the tRNA molecules enter the ribosome (the accommodation substep), which allows for tRNA to be accepted or rejected. His series of papers describes the conformational rearrangements that take place in the ribosome for this to happen properly. Elongation factors are proteins that assist protein synthesis, which can include the delivery of tRNA molecules to the ribosome. Whitford’s 2016 study in Nature Communications discovered that when elongation factor EF-Tu delivers the tRNA, if it slowly dissociates it may control the efficiency and accuracy of tRNA binding. In a follow-up study, Whitford showed how movements inside of the ribosome can also influence the accommodation process. Revealing this intricate relationship between ribosome and tRNA dynamics is gaining significant attention from the scientific community, where the recent study appeared on the cover of The Journal of Physical Chemistry. Together, these studies are beginning to reveal how ribosomal fluctuations and EF-Tu can come together to ensure efficient expression of our genome.
“It is rewarding to see how basic physical concepts can allow us to understand how these little engines work. It is also a very exciting time, since we are beginning to get our first glimpse of how this complicated machine functions,” Whitford said.
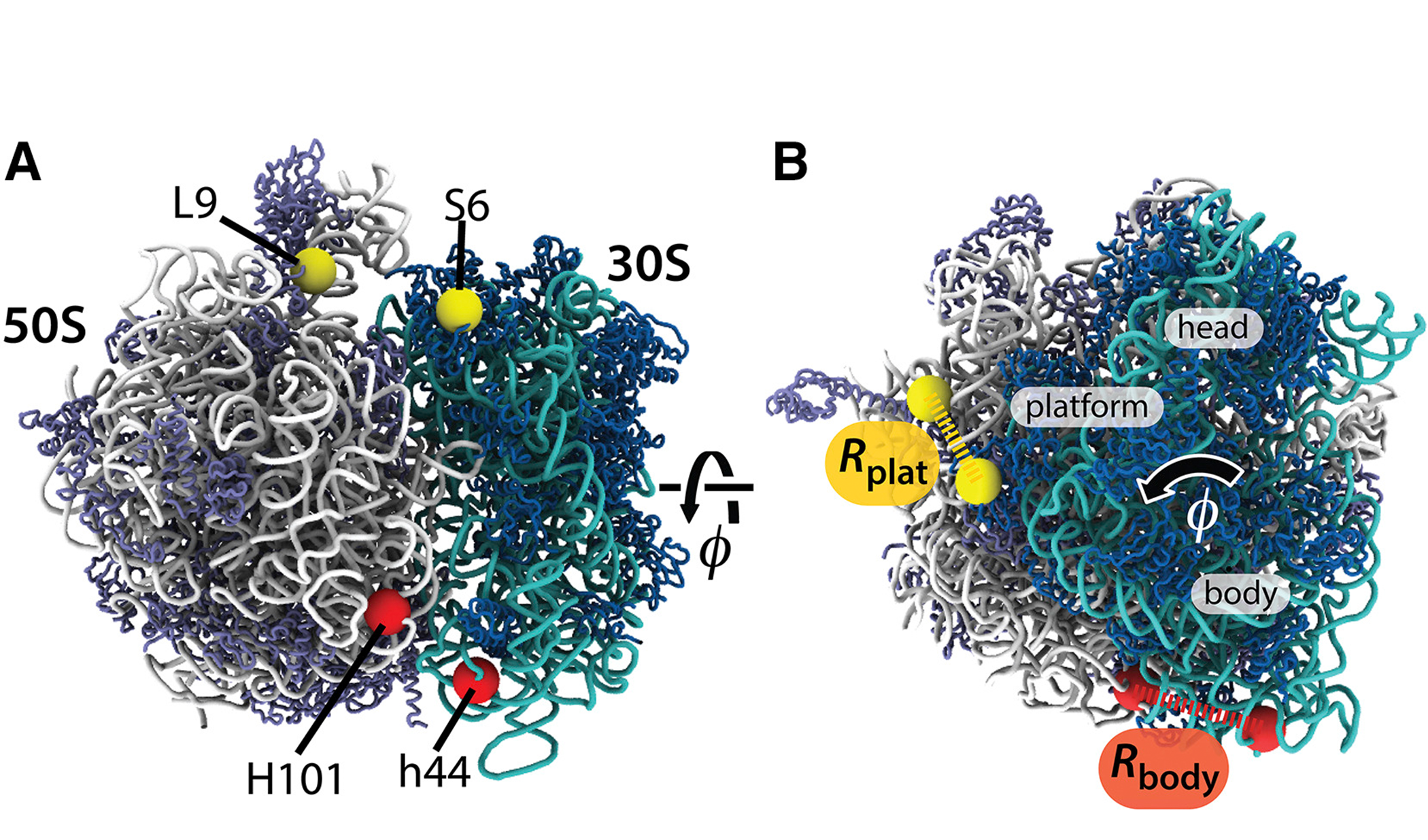
In a parallel line of inquiry, Whitford and his team have studied tRNA translocation, the process by which the ribosome resets itself before binding another tRNA molecule. This process involves global rearrangement of the ribosome, which make it particularly challenging to study. Whitford’s lab reported, in another paper published in Nature Communications, that there was a new, large-scale motion of about 20,000 atoms that moved together during this step. Without this movement, their work showed that the protein synthesis would likely stop.
“We’ve provided evidence of this new motion, and subsequent experimental studies have supported our finding. So, we’re very pleased that our theoretical tools are able to help guide the interpretation of experiments,” Whitford said.
Then in 2017, they took a more quantitative approach to measuring these large-scale motions. In their article published in Biophysical Journal, they showed how it may be possible to design new experiments that precisely monitor these rearrangements.
But Whitford won’t stop here. The ribosome undergoes a range of additional large-scale conformational motions, and so far, Whitford’s lab has looked at a few. Their work will continue to study the impacts that these motions have on the ribosome’s function and the cell as a whole. They also anticipate that future studies will elucidate how a range of different variables like temperatures or antibiotics affect the ribosome, as well as revealing the differences between human and bacterial ribosomes. There is no shortage of open questions that they can tackle next!