Inside each of the 37 trillion cells in the human body sits a blueprint for how to make every protein, structure, cell, bone, and organ we possess. Laid out, the DNA in each cell would be over 6 feet long, which doesn’t quite work when you have 37 trillion copies.
Our cells solve this problem by wrapping up our DNA like scrolls around structures called histones into packets called nucleosomes, packing it away in each cell’s nucleus for safekeeping. But, like scrolls, we can’t read our DNA when it is tightly wound around the histones, and we need to read our DNA to print copies of instructions that tell our cells how to perform their duties.
New research from Northeastern Professor Mark Williams and senior research scientist Micah McCauley has opened up this field, discovering the function of a protein that has long been a mystery.
The most common proteins in the nucleus of a cell are histones, the disks that DNA winds itself around. The second most common are a group called High Mobility Group proteins (HMGs).
“Besides histones, it’s the most abundant protein in the nucleus of eukaryotes, and we don’t know what it does,” McCauley said of HMGs.
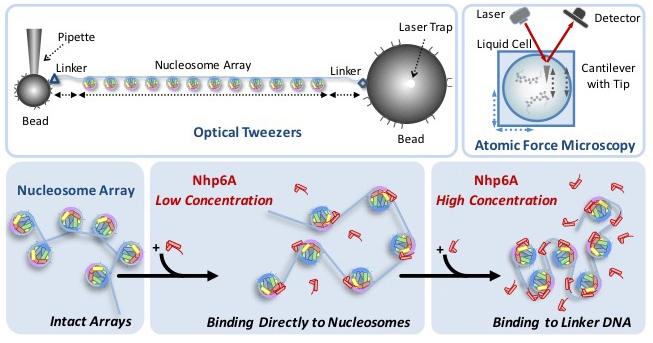
A depiction of DNA being unfurled with optical tweezers. Graphic provided by Mark Williams
There had been rumblings and guesses in the field about what they did, loosely that they had something to do with nucleosomes, these tight packets of DNA and proteins, but there hadn’t been a definitive test of their function.
Williams and McCauley wanted to change that. They believed that these proteins help the cell access its DNA, loosening the tight loops of DNA from their histones, giving a peek at the blueprint within.
To test their hypothesis, they drew on their colleagues across the nation. Dr. Karolin Luger of University of Colorado, the first to publish a reveal of the high-resolution structure of the nucleosome, and expert on DNA flexibility Dr. Jim Maher, Professor of Biochemistry and Molecular Biology at the Mayo Clinic.
Together, they designed an experiment based around the concept that these proteins should loosen DNA. To test that idea, they decided to pull the DNA apart. The researchers, along with then-graduate student Ran Huo, created strings of twelve nucleosomes and immersed them in liquids containing different amounts of HMG proteins. Using optical tweezers, an instrument that uses laser beams to apply force to single molecules, they pulled on the strings of nucleosomes and measured how much force was required to pop the nucleosomes off the string.
Williams and McCauley found that it was easier to pop the proteins off as the concentration of HMGs increased, suggesting that the HMGs loosen the tightly bound DNA when they bind to it. As concentrations got even higher, they found that the effect reversed; large amounts of HMGs leads to tightly bound DNA.
“If you add enough of these proteins they’ll start binding to the rest of the DNA and they’ll sort of cause it to collapse,” McCauley explained.
HMGs are different than most proteins in that they don’t need any additional energy to perform their function. They appear to be regulated purely by how much of it is present in the nucleus.
Their experimental design is naturally inspired, according to McCauley. “We’re learning how the cell makes DNA accessible and we’re doing it by imitating the process of pulling it apart, the process of accessing the packaged DNA.”
They confirmed their results using an Atomic Force Microscope, which allows them to visualize the process in still images. They had originally used the microscope just to see if their nucleosome strings looked right, but they realized there was more potential. “We thought, is there more information here that we could get?” McCauley explained that they came up with an analysis that allowed them to measure the distances between the nucleosomes, confirming their data from the optical tweezers.
“They complemented each other really nicely. Where they overlapped, they gave the same results, and we ran some simulations as well to try to mimic what we thought depositing [the strings] on a 2D surface would look like and that also gave very similar answers to what we saw with the optical tweezers.”
Even though this study was just published, Williams and McCauley are already looking ahead to the questions they will be asking next. “We know what HMG does, we don’t know how that is regulated, that’s the next step,” Williams says.
Because there’s no additional energy required for the proteins to function, it’s hard to say how the process gets started. That’s what the researchers will be working on next.
More questions remain about the nature of DNA, but the importance of this breakthrough as a stepping stone is recognized by Williams himself. “This is one small piece of that, but it does answer a big question about a very abundant protein, so it’s an important step overall.”