Imagine a novel antibiotic treatment that instantly kills bacteria and poses zero risks to humans. This therapy may be closer to reality than we once thought possible, thanks to the Whitford Research Group. Their recent publication in the esteemed journal Nature Communications highlights opportunities for the next generation of antibiotics through the study of the ribosome. The ribosome is a biomolecular machine responsible for producing the proteins that make up all living organisms. Every cell requires ribosomes to survive, including pathogens, like bacteria, that can be harmful to human health. Shutting down a ribosome means rendering a cell unable to survive – while bad news for destructive bacteria remains good news for scientists. A team at Northeastern University may have found a way to harness the functionality of the ribosome, elucidating a tremendous opportunity for the development of novel broad-spectrum antibiotics.
Imagine a novel antibiotic treatment that instantly kills bacteria and poses zero risks to humans. This therapy may be closer to reality than we once thought possible, thanks to the Whitford Research Group. Their recent publication in the esteemed journal Nature Communications highlights opportunities for the next generation of antibiotics through the study of the ribosome. The ribosome is a biomolecular machine responsible for producing the proteins that make up all living organisms. Every cell requires ribosomes to survive, including pathogens, like bacteria, that can be harmful to human health. Shutting down a ribosome means rendering a cell unable to survive – while bad news for destructive bacteria remains good news for scientists. A team at Northeastern University may have found a way to harness the functionality of the ribosome, elucidating a tremendous opportunity for the development of novel broad-spectrum antibiotics.
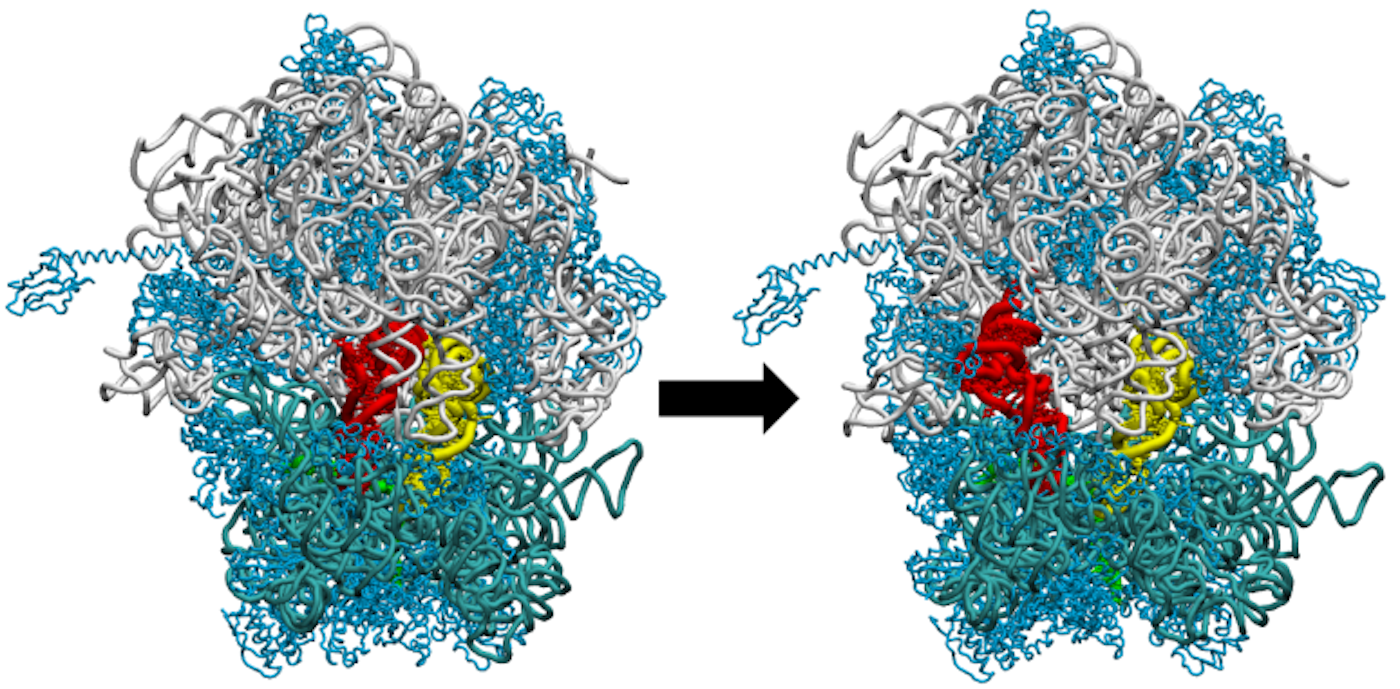
The team’s most recent published research, “A steric gate controls P/E hybrid-state formation of tRNA on the ribosome” was performed at the Massachusetts Green High Performance Computing Center (MHPCC). Funded by the National Science Foundation since 2014, Dr. Mariana Levi, Kelsey Walak, and Dr. Paul Whitford of Northeastern performed hundreds of simulations on structural elements of the ribosome using high performance computers. Their research focuses both on understanding the structure and function of the ribosome as well as finding key features that control its motion, with the ultimate goal of using them as experimental targets for potential antibiotics. “This is the first study where we actually have a strong signature of an antibiotic target,” said Dr. Whitford.
Harnessing the power of high-performance computing, a tool used for intensive computational tasks, the team developed a model that has identified a region on the ribosome unique to bacteria but absent from humans. Targeting this region pharmaceutically could kill bacterial cells without harming human ones. This finding is historic: “[Most previous studies] didn’t know where to look…it might take a thousand graduate student years to try out all of the possible regions that could be important…We’ve reduced this by maybe 95 percent,” says Dr Whitford. Working with the Research Computing Group at Northeastern, they went through hundreds of simulations to narrow down this target region using an in-house model developed by Dr. Levi. She credits the success of this model to the resources available to researchers for free at the MHPCC: “Our lab is our computer, so we really have to have a deeper understanding of what’s going on.”
This study is just the foundation for what is yet to come from this group. “We have something to say and add to the discussion,” says Dr. Levi. Their contributions extend not only to the community of ribosomal study but to those looking to leverage these innovative solutions to treat bacterial disease. “It’s much more than just computational simulation,” Dr. Levi adds, “This is just the beginning.”